AI-guided chemistry enables on-demand protein activation in living mice

Lisa Lock
scientific editor

Robert Egan
associate editor

To address the challenge of controlling protein activation in living animals for gain-of-function studies, researchers from Peking University led by Professors Chen Peng and Wang Chu recently developed CAGE-Proxvivo, a computer-aided proximal decaging strategy for on-demand protein activation and modulation of protein–protein interactions in living mice.
This work establishes a universal platform for time-resolved biological studies and on-demand therapeutic interventions in vivo. These findings were in Cell on May 27, 2025, under the title "Machine-learning-assisted universal protein activation in living mice."
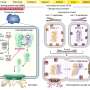
In-situ regulation and dynamic analysis of biologically active molecules are crucial for unveiling life processes and understanding disease mechanisms. The previously developed protein activation technology CAGE-Prox utilizes bioorthogonal cleavage reactions to install functional switches for various protein families, enabling regulation of protein function beyond specific activation sites.
This allows for universal protein activation with high spatial and temporal resolution, offering a novel strategy for dynamically analyzing protein mechanisms. However, this method relies on UV light, which has limited tissue penetration, thereby restricting its application in living animals.
The research teams employed the machine-learning-assisted evolution of aminoacyl-tRNA synthetase to successfully incorporate chemically caged amino acids into rationally designed "decaging sites," thereby transiently blocking target protein function. Functionality could then be restored in situ via a small-molecule-triggered bioorthogonal cleavage reaction.
Beyond the "active-pocket" decaging, CAGE-Proxvivo also enables precise control of protein–protein interactions, for example by a "gated" anti-CD3 antibody that permits chemically regulated T-cell recruitment and activation at tumor sites. It also allows tumor cell-specific activation of pyroptosis, which stimulates a robust anti-tumor immune response, showcasing its potential in immunotherapy.
This method demonstrates broad applicability, from activating proteins of interest to cell-type-specific modulation of distinct phenotypes in living systems.
More information: Xin Wang et al, Machine-learning-assisted universal protein activation in living mice, Cell (2025).
Journal information: Cell
Provided by Peking University