Illustration of the new analysis method. The tractors represent so-called transcription factors: proteins that bind to genetic switches to activate or deactivate genes. The method compares the genetic material of two parents with different traits (illustrated by the different size), within a hybrid plant. This comparison makes it possible to determine if a change in the switch sequence (orange boxes) results in stronger or weaker binding of the transcription factors, thereby altering the traits. Credit: HHU/Andi Kur (under license BY-NC-SA)
Natural genetic variation in the genome ensures biodiversity and drives evolution. However, as natural evolutionary processes require millennia, we cannot wait for them to adapt crop plants to the rapidly changing climatic conditions. To safeguard global food security, researchers must accelerate the identification of appropriate natural DNA variants to improve crop plant performance under stress conditions.
A research team headed by Dr. Thomas Hartwig and Dr. Julia Engelhorn from the Institute for Molecular Â鶹ÒùÔºiology at HHU and MPIPZ now presents a new, efficient method for mapping the genetic "switches" of plants in a current in Nature Genetics. Not actually genes themselves, these small sections of the genome determine when, where and to what extent a gene is active. They are comparable with a dimmer switch regulating the brightness of a lamp.
While research so far has largely focused on the genes themselves, the new study demonstrates that key differences between plants—e.g. variation in size, or resistance to diseases or stress situations—are often not determined by the genes, but rather by these regulatory switches. Traditionally, however, it is not only difficult to locate these regions precisely, but also to determine which changes play the decisive role. This is now changing thanks to a new, scalable mapping method developed within the framework of the project.
The research team analyzed 25 different maize hybrids, i.e. crossbreeds of different maize varieties, identifying over 200,000 regions in the genome where natural variations influence regulatory switches.
Engelhorn, lead author of the study, says, "Although these regulatory switches make up less than 1% of the genome, the variations often explain a substantial share of heritable trait differences—sometimes exceeding half."
Hartwig, corresponding author of the study, comments, "Understanding how these regulatory switches operate provides a powerful new tool to enhance both crop resilience and yield—laying the foundation for smarter breeding processes in the future."
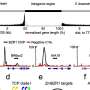
Quantitative cis-element occupancy analysis in F1 hybrids. Credit: Nature Genetics (2025). DOI: 10.1038/s41588-025-02246-7
The researchers applied their method specifically to traits, which play a role in drought stress, identifying over 3,500 individual regulatory switches and the associated genes via which the plants respond to water-limited conditions.
Engelhorn says, "Our approach allows direct comparison of the differences in switch variants inherited via the maternal and paternal lines in a single experiment. We can thus offer the maize research community a resource of over 3,500 drought-linked regulatory sites—opening up new possibilities to fine-tune gene expression for enhanced robustness."
Hartwig adds, "The precision of this mapping enables us to learn from the natural differences in the switches how they work, which in turn enables targeted manipulation of the switches to develop plants with improved traits."
This research was realized in collaboration with a team from the University of California in Davis, in which Dr. Samantha Snodgrass is a member. The co-author of the study emphasizes the change in perspective accompanying the approach: "Despite decades of successful research, much of the genome—the parts outside the genes—remains a black box. This new method pulls back the curtain and enables us to identify the function of these non-coding areas, providing biologists and breeders with new, precise targets for new research and development approaches."
The study was conducted within the CEPLAS Cluster of Excellence on Plant Sciences at HHU and MPIPZ.
More information: Julia Engelhorn et al, Genetic variation at transcription factor binding sites largely explains phenotypic heritability in maize, Nature Genetics (2025).
Journal information: Nature Genetics
Provided by Heinrich-Heine University Duesseldorf