Automated tool enables rapid, large-scale profiling of disease-linked RNA modifications

Gaby Clark
scientific editor

Robert Egan
associate editor

Researchers have developed a powerful tool capable of scanning thousands of biological samples to detect transfer ribonucleic acid (tRNA) modifications—tiny chemical changes to RNA molecules that help control how cells grow, adapt to stress and respond to diseases such as cancer and antibiotic‑resistant infections. This tool opens up new possibilities for science, health care and industry—from accelerating disease research and enabling more precise diagnostics, to guiding the development of more effective medical treatments for diseases such as cancer and antibiotic‑resistant infections.
The research was led by SMART AMR, in collaboration with Nanyang Technological University (NTU Singapore), University of Florida, University at Albany, Lodz University of Technology and MIT.
Addressing current limitations in RNA modification profiling
Cancer and infectious diseases are complicated health conditions in which cells are forced to function abnormally by mutations in their genetic material or by instructions from an invading microorganism. The SMART-led research team is among the world's leaders in understanding how the epitranscriptome—the over 170 different chemical modifications of all forms of RNA—controls growth of normal cells and how cells respond to stressful changes in the environment, such as loss of nutrients or exposure to toxic chemicals. The researchers are also studying how this system is corrupted in cancer or exploited by viruses, bacteria and parasites in infectious diseases.
Current molecular methods used to study the expansive epitranscriptome and all of the thousands of different types of modified RNA are often slow, labor‑intensive, costly and involve hazardous chemicals which limit research capacity and speed.
To solve this problem, the SMART team developed a new tool that enables fast, automated profiling of tRNA modifications—molecular changes that regulate how cells survive, adapt to stress and respond to disease. This capability allows scientists to map cell regulatory networks, discover novel enzymes and link molecular patterns to disease mechanisms, paving the way for better drug discovery and development, and more accurate disease diagnostics.
Unlocking the complexity of RNA modifications
SMART's research, recently in Nucleic Acids Research, titled "tRNA modification profiling reveals epitranscriptome regulatory networks in Pseudomonas aeruginosa", shows that the tool has already enabled the discovery of previously unknown RNA‑modifying enzymes and the mapping of complex gene regulatory networks. These networks are crucial for cellular adaptation to stress and disease, providing important insights into how RNA modifications control bacterial survival mechanisms.
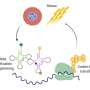
Using robotic liquid handlers, researchers extracted tRNA from more than 5,700 genetically modified strains of Pseudomonas aeruginosa, a bacterium that causes infections such as pneumonia, urinary tract infections, bloodstream infections and wound infections. Samples were enzymatically digested and analyzed by liquid chromatography‑tandem mass spectrometry (LC‑MS/MS), a technique that separates molecules based on their physical properties and identifies them with high precision and sensitivity.
As part of the study, the process generated over 200,000 data points in a high‑resolution approach that revealed new tRNA‑modifying enzymes and simplified gene networks controlling how cells respond and adapt to stress. For example, the data revealed that the methylthiotransferase MiaB, one of the enzymes responsible for tRNA modification ms2i6A, was found to be sensitive to the availability of iron and sulfur and to metabolic changes when oxygen is low. Discoveries like this highlight how cells respond to environmental stresses, and could lead to future development of therapies or diagnostics.
SMART's automated system was specially designed to profile tRNA modifications across thousands of samples rapidly and safely. Unlike traditional methods—which are costly, labor‑intensive and use toxic solvents such as phenol and chloroform—this tool integrates robotics to automate sample preparation and analysis, eliminating the need for hazardous chemical handling and reducing costs. This advancement increases safety, throughput and affordability, enabling routine large‑scale use in research and clinical labs.

A faster and automated way to study RNA
As the first system capable of quantitative, system‑wide profiling of tRNA modifications at this scale, the tool provides a unique and comprehensive view of the epitranscriptome—the complete set of RNA chemical modifications within cells. This capability allows researchers to validate hypotheses about RNA modifications, uncover novel biology and identify promising molecular targets for developing new therapies.
"This pioneering tool marks a transformative advance in decoding the complex language of RNA modifications that regulate cellular responses. Leveraging AMR's expertise in mass spectrometry and RNA epitranscriptomics, our research uncovers new methods to detect complex gene networks critical for understanding and treating cancer as well as antibiotic‑resistant infections.
"By enabling rapid, large‑scale analysis, the tool accelerates both fundamental scientific discovery and the development of targeted diagnostics and therapies that will address urgent global health challenges," said Prof Peter Dedon, Co‑lead Principal Investigator (PI) at SMART AMR, Professor of Biological Engineering at MIT and corresponding author of the paper.
Accelerating research, industry and health care applications
This versatile tool has broad applications across scientific research, industry and health care. It enables large‑scale studies of gene regulation, RNA biology and cellular responses to environmental and therapeutic challenges. The pharmaceutical and biotech industry can harness it for drug discovery and biomarker screening, efficiently evaluating how potential drugs affect RNA modifications and cellular behavior. This aids the development of targeted therapies and personalized medical treatments.
"This is the first tool that can rapidly and quantitatively profile RNA modifications across thousands of samples. It has not only allowed us to discover new RNA‑modifying enzymes and gene networks, but also opens the door to identifying biomarkers and therapeutic targets for diseases such as cancer and antibiotic‑resistant infections. For the first time, large‑scale epitranscriptomic analysis is practical and accessible," said Dr. Jingjing Sun, Research Scientist at SMART AMR and first author of the paper.
Looking ahead: Advancing clinical and pharmaceutical applications
Moving forward, SMART AMR plans to expand the tool's capabilities to analyze RNA modifications in human cells and tissues, moving beyond microbial models to deepen understanding of disease mechanisms in humans.
Future efforts will focus on integrating the platform into clinical research to accelerate the discovery of biomarkers and therapeutic targets. The translation of the technology into an epitranscriptome‑wide analysis tool that can be used in pharmaceutical and health care settings, will drive the development of more effective and personalized treatments.
More information: Jingjing Sun et al, tRNA modification profiling reveals epitranscriptome regulatory networks in Pseudomonas aeruginosa, Nucleic Acids Research (2025).
Journal information: Nucleic Acids Research
Provided by Singapore-MIT Alliance for Research and Technology (SMART)