Spotting stealth multiplets for better single-cell experiment design

Lisa Lock
scientific editor

Robert Egan
associate editor

Single-cell RNA sequencing (scRNA-seq) has revolutionized modern biology, allowing scientists to study the expression of genes from thousands of individual cells at the same time. This technique helps researchers identify different cell populations, study cellular heterogeneity, and understand gene expression dynamics in complex tissues.
In a recent study, researchers from EMBL Barcelona provided new insights into some of the technical limitations of the process, which could help scientists better address these risks while designing scRNA-seq experiments.
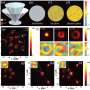
A common scRNA-seq method utilizes a microfluidic device generating millions of "microdroplets." Each cell is captured in a single microdroplet and then subjected to biochemical profiling. However, on random occasions, multiple cells are captured at the same time in a microdroplet and wrongly recognized as a single cell (called a "multiplet").
This hidden artifact can distort data and mislead the interpretation of experiments. One way to reduce the risk of multiplets is "sample multiplexing," a technique where samples are labeled with different molecular barcodes, and multiplets can be distinguished by the presence of multiple barcodes in the same microdroplet. Although this strategy has improved cost-efficiency and experimental design, it was unclear whether it still retained a risk of hidden multiplets.
In the new study, Fumio Nakaki, Postdoctoral Fellow at EMBL Barcelona's Sharpe Group, has quantitatively evaluated the risk of having undetected (or "stealth") multiplets during sample multiplexing, both in theory and in practice. The work has been in BMC Genomics.
"Our goal is to provide researchers with a practical reference point," said Nakaki. "Sample multiplexing is widely used, but its limitations are often overlooked. By clarifying how and when multiplets arise under multiplexed conditions, we hope this work will support better study design in experiments and data interpretation."
To assess how sample multiplexing influences multiplet detection, Nakaki combined theoretical modeling with experimental benchmarking and used publicly available single-cell datasets. He derived mathematical expressions to estimate expected multiplet rates under various input conditions and validated these predictions against real data where individual samples had been uniquely barcoded.
By comparing different multiplexing configurations, Nakaki showed that while sample multiplexing helps identify some types of multiplets, at least one multiplet type still remains, especially in conditions where the sample labeling was insufficient or the choice of analysis tools was inadequate.
Their analysis provides practical guidance on how to balance cell input, sample number, and desired specificity, highlighting that even with multiplexing, experimental parameters must be carefully optimized to maintain the integrity of single-cell data.
More information: Fumio Nakaki et al, Probability of stealth multiplets in sample-multiplexing for droplet-based single-cell analysis, BMC Genomics (2025).
Journal information: BMC Genomics
Provided by European Molecular Biology Laboratory