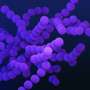
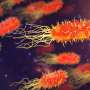
Continuum model versus agent-based model for an initially uniform bacterial population responding to a decaying pulse of chemo effector. Credit: Philosophical Transactions of the Royal Society A: Mathematical, Â鶹ÒùÔºical and Engineering Sciences (2025). DOI: 10.1098/rsta.2024.0261
Newcastle scientists have developed an efficient new model to track bacterial responses to toxic chemicals and nutrients.
The model helps explain how bacteria react in places like oceans or soil, where chemical signals, including nutrients or pollutants, can change quickly. Better understanding of these population dynamics is essential to safeguard ecosystems that rely on bacteria and are becoming more vulnerable due to challenges from pollution and climate change.
Examples of bacteria navigating time-changing chemical environments in nature include pathogenic bacteria in the human body, where chemical levels can change rapidly over time and bacteria need to be able to swim away from chemicals that are toxic to them, such as antibiotics (negative chemotaxis). In soils, bacteria also need to seek out sugars exuded by plant roots (positive chemotaxis), which change in time as the root only exudes sugars at certain times of day.
While older models only looked at how bacteria respond to chemical changes in space, the new research shows that changes over time, such as chemicals appearing or disappearing, can also affect how bacteria move.
Published in the journal Philosophical Transactions of the Royal Society A, the allows the description of populations of bacteria at a lower computational cost, and thus reduced carbon footprint, than previous models.
Study lead author, Jason Bains, Ph.D. researcher at Newcastle University's School of Mathematics, Statistics and Â鶹ÒùÔºics, explains, "Previous models which modeled bacteria individually can model these dynamics, but at very high computational cost and reduced population numbers, our model can achieve this with a much lower computational cost by modeling the population as a whole.
"This is important because these models need to be accessible to all researchers working on the front line of microbial science who may not always have access to large computational resources."
Principal investigator of the study, Dr. Otti Croze, added, "Bacterial communities in the environment comprise enormous numbers of individuals. Simulating even a fraction of these individuals would be impossible for even the most powerful computers. Our model allows large chemotactic bacterial populations to be simulated at low computational cost, which can help understand microbial communities, from the soil, stimulating sustainable agriculture, to the human body, helping to tackle antimicrobial resistance."
More information: Jason Singh Bains et al, Drift velocity of bacterial chemotaxis in dynamic chemical environments, Philosophical Transactions of the Royal Society A: Mathematical, Â鶹ÒùÔºical and Engineering Sciences (2025).
Journal information: Philosophical Transactions of the Royal Society A
Provided by Newcastle University