How enzymes 'dance' while they work, and why that's important

Sadie Harley
scientific editor

Robert Egan
associate editor

Researchers from Tokyo Metropolitan University have developed a new structure determination method using nuclear magnetic resonance (NMR) spectroscopy which shows how different parts of complex molecular machinery like enzymes move while they help catalyze reactions. The research is in the Journal of the American Chemical Society.
Focusing on an enzyme in yeast, they demonstrate how contrasts in atomic scale motions impact their function. The method promises unprecedented access to the mechanisms by which biomolecules work, and how they relate to illnesses.
Enzymes are indispensable to the function of all biological organisms, including humans. While snapshots captured using X-ray or cryo-electron microscopy have revealed their intricate molecular structure, they are a constant blur of motion when they are at work.

Their atomic scale structure is constantly changing, capturing other biomolecules and helping them react in a carefully choreographed sequence. While clearly important, these rapid motions at the nanometer scale are difficult to capture.
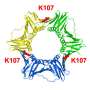
By integrating a combination of different approaches, the researchers led by Associate Professor Teppei Ikeya from Tokyo Metropolitan University, successfully captured an accurate "ensemble structure" of a reacting enzyme. Essentially, ensemble structure is the collection of all states a macromolecule can take, and how likely it is to take each.
The team demonstrated their method with yeast ubiquitin hydrolase 1 (YUH1), an enzyme in yeast that helps it recycle ubiquitin, a protein that regulates various intracellular events.
YUH1 has an analog in humans called ubiquitin C-terminal hydrolase (UCHL1), known to be implicated in Parkinson's and Alzheimer's diseases. By integrating multiple analytical methods using NMR spectroscopy, they were able to create an ensemble map for YUH1 for dynamics over the millisecond time scale.

They discovered that two parts of the enzyme near the active part, where proteins are bound, showed strikingly large movements, a "crossover loop" structure and the N-terminus, one end of the protein-capturing structure of the enzyme.
The N-terminus was found to move in and out of the loop, going through a whole range of states before finally capturing a target protein like a lasso before acting as a "gating lid," keeping it in place. This new mechanism was supported by the way in which mutant versions with incomplete "gating lids" failed to show the same enzymatic activity.
The team's findings reveal how the dynamic nature of enzymes plays an important role in how they function.
The method can be applied to a vast array of biological structures in their natural environment, promising a new approach for scientists to access underlying mechanisms and potential pathologies.
More information: Mayu Okada et al, Multistate Structure Determination and Dynamics Analysis Reveals a Unique Ubiquitin-Recognition Mechanism in Ubiquitin C-terminal Hydrolase, Journal of the American Chemical Society (2025).
Journal information: Journal of the American Chemical Society
Provided by Tokyo Metropolitan University