New measurement method visualizes chemical signals of individual cells

Lisa Lock
scientific editor

Robert Egan
associate editor

Diagnosing cancer and selecting the appropriate therapy depend crucially on how well experts understand the processes in tumors at the microscopic level. Central to this is understanding how cells in tissues communicate and what chemical signals are involved. Innovative methods that make changes visible at the single-cell level can lead to faster and more precise diagnoses and more targeted therapies, thereby improving treatment outcomes.
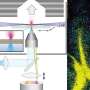
A research team from the Institute of Hygiene at the University of Münster has developed a new measurement method that combines fluorescence microscopy directly with MALDI mass spectrometry imaging for the first time. MALDI, or "matrix-assisted laser desorption/ionization," allows researchers to determine the chemical profiles of individual neighboring cells in tissue samples on the same section grid and with a spatial resolution of about one thousandth of a millimeter.
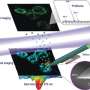
With this method, the team was able to visualize previously hidden, different metabolic patterns between immediately neighboring cells in tumor tissue. The results have been in the journal Nature Communications.
"For the first time, we are able to identify cell types based on fluorescence and match them with their chemical signature in the tissue context. This allows us to detect chemical differences and interactions at the single-cell level," says Dr. Alexander Potthoff, first author of the study. This is particularly relevant because the interaction between cancer cells, surrounding cells, and invading immune cells in tumors often determines whether the disease remains localized or begins to spread.
The MALDI mass spectrometry imaging technique uses a laser to release molecules from the tissue and measure their mass. This provides information on numerous metabolites and cell wall components. However, since a mass spectrometer only detects electrically charged particles, a second laser can be used for post-ionization. This so-called MALDI-2 technique significantly increases the detection sensitivity for many important molecule classes.
The method now being presented combines two technical improvements: first, the use of an inverse irradiation geometry, also known as transmission mode, which increases spatial resolution. Second, the integration of a fluorescence microscope directly into the mass spectrometer used.
With an optimized sample preparation, this allows researchers to directly couple fluorescence-based measurements, e.g., on a protein basis, with mass spectrometric analysis of the metabolome and lipidome—on exactly the same tissue section.
The idea of increasing detection sensitivity by using a second laser was presented several years ago by the Münster research team. The transmission mode had also been described previously. The combination of these components with a directly integrated fluorescence microscope and the adaptation of sample preparation are new.
"The combined method could support numerous established techniques in fluorescence microscopy. Researchers in basic research, for example in cell biology, immunology, and tumor biology, are likely to benefit in particular," explains Dr. Jens Soltwisch. From a clinical perspective, therapy decisions in the future could be supported by a complementary, rapid analysis of biopsies.
According to Prof Dr. Klaus Dreisewerd, there is even more potential: "With further technical improvements, the spatial resolution could advance to the range of a few hundred nanometers, so that even the chemical composition of individual cell organelles such as intracellular lipid droplets, vesicles or synapses could be examined."
In the long term, such findings will help scientists develop new active substances and make health care systems more efficient.
More information: Alexander Potthoff et al, Spatial biology using single-cell mass spectrometry imaging and integrated microscopy, Nature Communications (2025).
Journal information: Nature Communications
Provided by University of Münster