Gene-reading enzyme, inhibitor protein interaction analysis provides surprising insights
Within the cells, the RNA polymerase (RNAP) protein complex clutches DNA like a crab claw, scanning across gene-coding regions and transcribing these sequences into the messenger RNA molecules that will ultimately provide the blueprint for protein production.
This process can be impaired or assisted through interactions with proteins known as transcription factors, but understanding how these factors influence RNAP function can pose a serious challenge for structural biologists. “It is very difficult to crystallize RNAP, which is an unusually large enzyme,” says Shigeyuki Yokoyama, director of the RIKEN Systems and Structural Biology Center in Yokohama. “In particular, no crystal structures of bacterial RNAP-transcription factor complexes have ever been reported.” Recently, however, Yokoyama and colleagues successfully obtained a crystal structure that captures RNAP in the midst of transcription while bound to Gre factor homologue 1 (Gfh1), a transcription factor from the bacterium Thermus thermophilus.
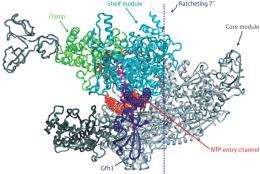
RNAP consists of several discrete modules connected by flexible linker regions, with most of the enzymatic machinery residing in the ‘shelf’ and ‘core’ modules that serve as the main body of the RNAP ‘claw’. In their structure, the researchers uncovered a never-before-seen arrangement of the RNAP modules, where some sort of ‘ratcheting’ action has created notable displacement between the shelf and core relative to its normal structure.
In fact, the binding of Gfh1 appears to lock RNAP into this configuration. This transcription factor—a known inhibitor—inserts itself into a channel on the complex that normally accepts nucleotides for addition onto newly synthesized RNA molecules (Fig. 1). However, such insertion would be impossible with the normal RNAP complex, where the channel is too narrow. This suggests that RNAP executes this unexpected ratcheting motion as part of its normal behavior, which in turn leaves it vulnerable to Gfh1 inhibition. “This conformational change was most surprising,” says Yokoyama. “It was simply impossible to predict this before the structure of RNAP-Gfh1 was solved.”
In subsequent biochemical experiments, he and his colleagues managed to essentially catch RNAP in the act of ratcheting, providing further evidence that this behavior occurs spontaneously in nature and is likely to contribute directly to the enzyme’s transcriptional activity. “We hypothesize that RNAP uses this ratcheted state to slide along DNA chains as an intermediate step in the course of normal transcription,” says Yokoyama. “This state may also be used an intermediate for transcriptional termination, in which the [RNA] dissociates from the RNAP.” He adds that validating these and other hypotheses will be top priorities for future experimental efforts.
More information: Tagami, S., et al. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature 468, 978–982 (2010).
Provided by RIKEN