June 14, 2019 feature
Raising fluid walls around living cells

Thamarasee Jeewandara
contributing writer
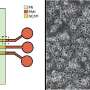
Cell culture plates that are in everyday use in biology can be effectively transformed into microfluidic devices, opening paths for biologists to miniaturize cell-based workflows. In a recent report, Ph.D. researcher Cristian Soitu and co-workers in the departments of Engineering Science and Pathology at the University of Oxford, Oxford, U.K., described a simple method to create microfluidic arrangements around cells. In the study, the cells were already growing on standard surfaces, when the scientists used the interface between the immiscible fluid media in the container as a building material.
They re-purposed the conventional cell culture dishes into sophisticated microfluidic devices on demand by reshaping fluid structures around living cells. Soitu describes the new fluid-shaping technique built by his research team as "fluid structures for those cells with fear of commitment when choosing a home—they can be easily removed and new ones (with a different geometry) built in place." The research is now published on Science Advances
The researchers demonstrated the method using workflows involving cell cloning; selective cloning of a specific clone from among others in a dish; drug treatments; and wound healing. The research work demonstrated a versatile approach, coupled to biologically friendly features to promote the microfluidics technology among biologists. Microfluidics-based approaches have gained popularity in , although their uptake in mainstream biology remains slow due to a , including:
- Material incompatibility for cell growth
- Microfluidic architectures that are enclosed and inaccessible
- Predetermined geometries that cannot be reconfigured during experiments—causing cost of manufacture and operation
- Workflows that do not align with pre-existing techniques developed by biologists.
In the past, scientists created 3-D constructs with fluid walls at the nanoscale, although their . In the present work, therefore, Soitu et al. developed a method to make arrays of isolated microfluidic chambers on virgin petri dishes to accommodate major workflows in cell biology. Possible examples include cell feeding and transfer, cloning, cryopreservation, fixation and immunolabeling, cell lysis and (RT-PCR) and CRISPR-Cas9 gene editing. In previous experiments of such workflows scientists added the .
In the present work, the researchers created a variety of microfluidic arrangements on standard petri dishes containing adherent cells and reconfigured them in real-time. They isolated and retrieved cell clones to perform proof-of-concept drug tests and wound-healing assays and introduced the new technique to create and reconfigure microfluidic circuits on petri dishes while cells grew and divided, with many potential applications in mainstream biology.
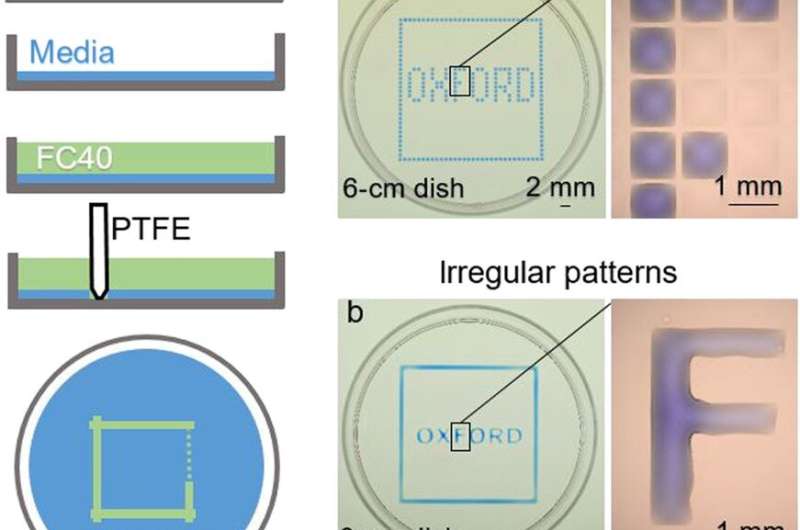
The new technique and the proof-of-concept experiments
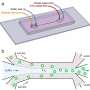
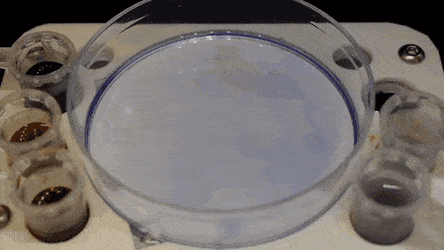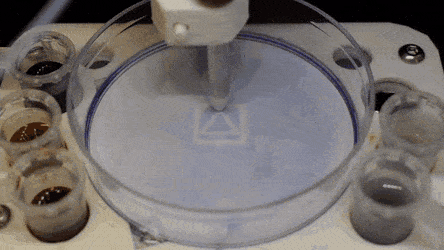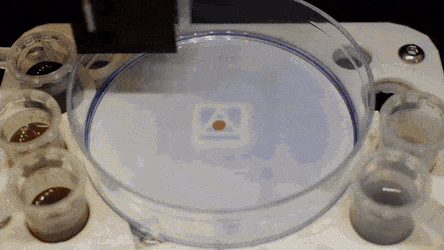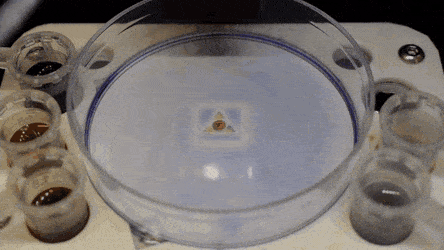
In the experiments that followed, the researchers first covered the bottom of a petri dish with tissue culture medium and removed most of the medium to form a thin film covering the polystyrene substrate. They overlaid the thin film with an immiscible fluorocarbon (FC40) to prevent evaporation and as a barrier against external contaminants to maintain sterility of the medium. Then using a Teflon tip, the researchers contacted the bottom of the dish, displacing the aqueous phase to form microfluidic arrangements in the shape of interest—in this instance, a square. Using the technique, the researchers brought the open microfluidic platform's advantages to standard cell culture-ware.
Soitu et al. shaped the aqueous phase to create a grid with as previously demonstrated by the same team, and viewed them with selective dyes in selective chambers. For instance, the peripheral chambers received a blue dye (forming a blue square) and those in the interior formed the word "OXFORD."
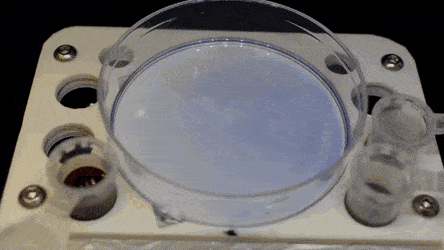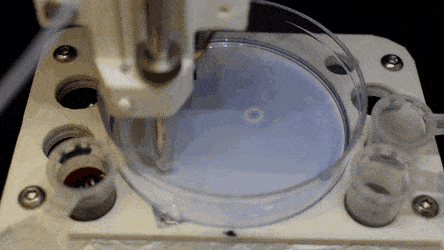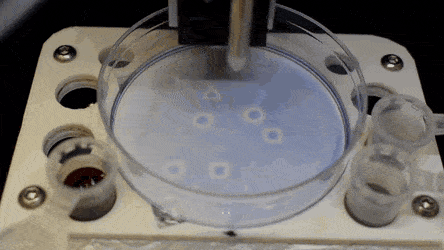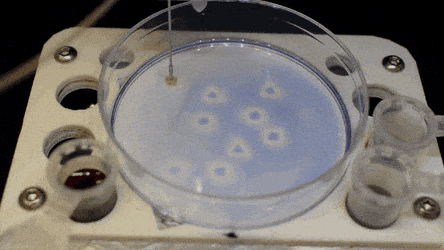
The researchers "printed" a circle within a triangle within a square and used microliters of three dyes to separately view the three shapes; where FC40 prevented the dyes from mixing. The results showed capability to build and destroy FC40 walls to effectively confine the liquids in any desired 2-D shape.
After the preliminary proof-of-concept results, Soitu et al. generated arrays of chambers to recapitulate the cloning of (NM18), for which they initially created grids, followed by cell addition thereafter. The researchers first allowed the cells to grow freely surrounded by the FC40 wall permeable to both O2 and CO2, and then by growing single cells into clones before surrounding them with fluid walls of different shapes.
They showed that fluid walls with different 2-D footprints could be built easily around living cells, as long as the colonies remained isolated from each other during subsequent treatment or retrieval. that grew cells within confined, pre-patterned surfaces required surface treatment prior to cell adhesion—contributing to the notable exception in the present technique.
Applications in clone picking and drug testing
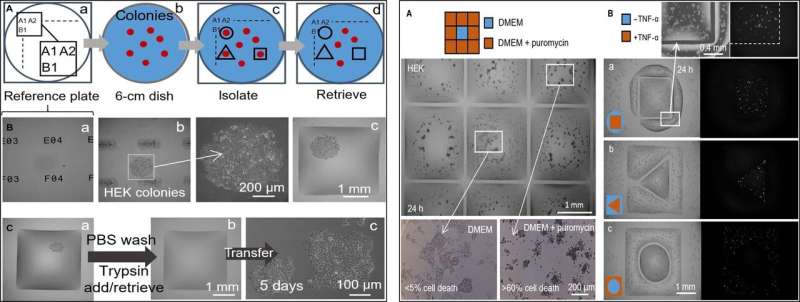
In the next step, the researchers created a reference plate on which they placed a dish containing live cell colonies of interest to isolate cell clones of interest from others by printing fluid walls around them. On isolation, they could pick the colonies, recover the cells and grow them conventionally to multiply as expected. Since the fluid walls could effectively confine the liquids, Soitu et al. tested their efficiency by adding – a small molecule translator inhibitor that kills mammalian cells.
In the drug screening experimental setup, they allowed the central chamber to receive growth medium alone, while the drug was delivered to the surrounding chambers in a high lethal dose, to show the efficacy of FC40 separation when only the cell lines in the central chamber survived. In a second example, Soitu et al. exploited the property of a genetically modified to encode a green fluorescent promoter gene. Which switched on in the presence of to fluoresce green. The fluid walls formed effective barriers to drug exposure, verifying the technique's drug screening potential.
Applications in wound healing
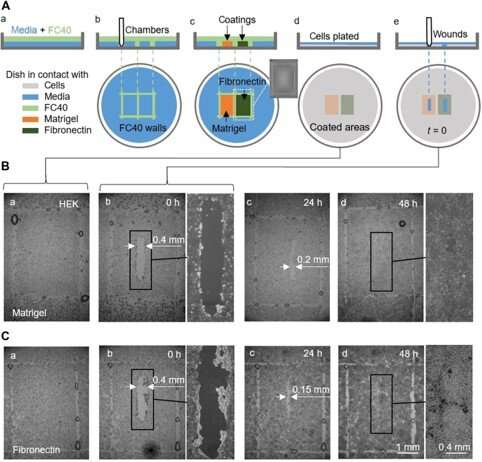
They also completed proof-of-principle wound healing assays by using a single dish coated in two different ways, to monitor two wound healing conditions. For this, the researchers used —a gelatinous protein secreted by sarcoma cells —a glycoprotein of the extracellular matrix that improved wound healing. They added HEK cells that formed a monolayer in the dishes and created a "wound" by dragging the Teflon tip across the monolayer when cells migrated into the wounds at slightly different rates. Although in this workflow Soitu et al. pre-coated the surface before plating cells, they could also modify the coating technique for its addition after the cells began migrating into the newly formed wounds to promote healing.
In this way, Cristian Soitu and co-workers developed a flexible, microfluidic platform to miniaturize workflows in cell biology. They extended the technique in the present work to form microfluidic arrangements around pre-plated adherent cells followed by a variety of proof-of-principle assays on cell cloning, drug screening and wound healing. The platform has many advantages and can replace the conventional mode of pre-engineered microfluidic devices as a flexible and customizable alternative. The new microfluidic arrangements are cost-effective contributing to and can be reconfigured in real-time during an experiment for added versatility. The researchers note limitations of the technique, including 2-D restricted arrangements and the fragility of fluid walls compared to solid walls. Soitu et al. hope to optimize and combine these features and advantages to provide a new platform for mainstream biologists to explore the power of microfluidics.
Written for you by our author —this article is the result of careful human work. We rely on readers like you to keep independent science journalism alive. If this reporting matters to you, please consider a (especially monthly). You'll get an ad-free account as a thank-you.
More information: Cristian Soitu et al. Raising fluid walls around living cells, Science Advances (2019).
Eric K. Sackmann et al. The present and future role of microfluidics in biomedical research, Nature (2014).
Cristian Soitu et al. Microfluidic chambers using fluid walls for cell biology, Proceedings of the National Academy of Sciences (2018).
Edmond J. Walsh et al. Microfluidics with fluid walls, Nature Communications (2017).
Journal information: Science Advances , Nature , Proceedings of the National Academy of Sciences , Nature Communications
© 2019 Science X Network